Atac-seq Which Index to Use With One Sample
In the interest of time and available compute power We will work on a single chromosome in this tutorial. Keep the mixture at room temperature 21C23C before use in step 12.
Atac Seq With Bioconductor Atacseqworkshop
Pipette Tip Recommendations for 10x Genomics Single Cell Protocols.

. Kit Performance Correlates with Published Data. Experimentally determined number of additional cycles. To promote delivery of high-quality data for cyropreserved cells we ask customers to provide 3 tubes per sample with 100000-200000 cells and 90 viability for ATAC-Seq library preparation.
You will see this file has a copy of the parameters in atac-exampleyaml for each sample. As performed for single cell RNA-seq single nuclei prep will be check first for quality and concentration and 500-10000 nuclei can be targeted in each sample. - Use 50 bp paired-end 50PE sequencing.
Read 1 Read 2 sample ID. SRR891269 see in RawDataATAC folder for fastq and bam files. Please do not send more than 100000 cells.
Active Motif will accept the following sample types for this service. Run the analysis This will run the analysis on a local machine using 16 cores. If your sample has less than 100000 cells a member of our team can discuss options for your project.
An example is below. Therefore the user simply needs to. PCR amplification of the ATAC-seq library Done in two stages.
Reagents Workflow Data Overview. If using primary cell cultures treating cells with compounds that may induce cell death or cells that. Active Motif ATAC-Seq Sample Preparation Protocol Rev D_092020 ATAC-Seq Sample Preparation.
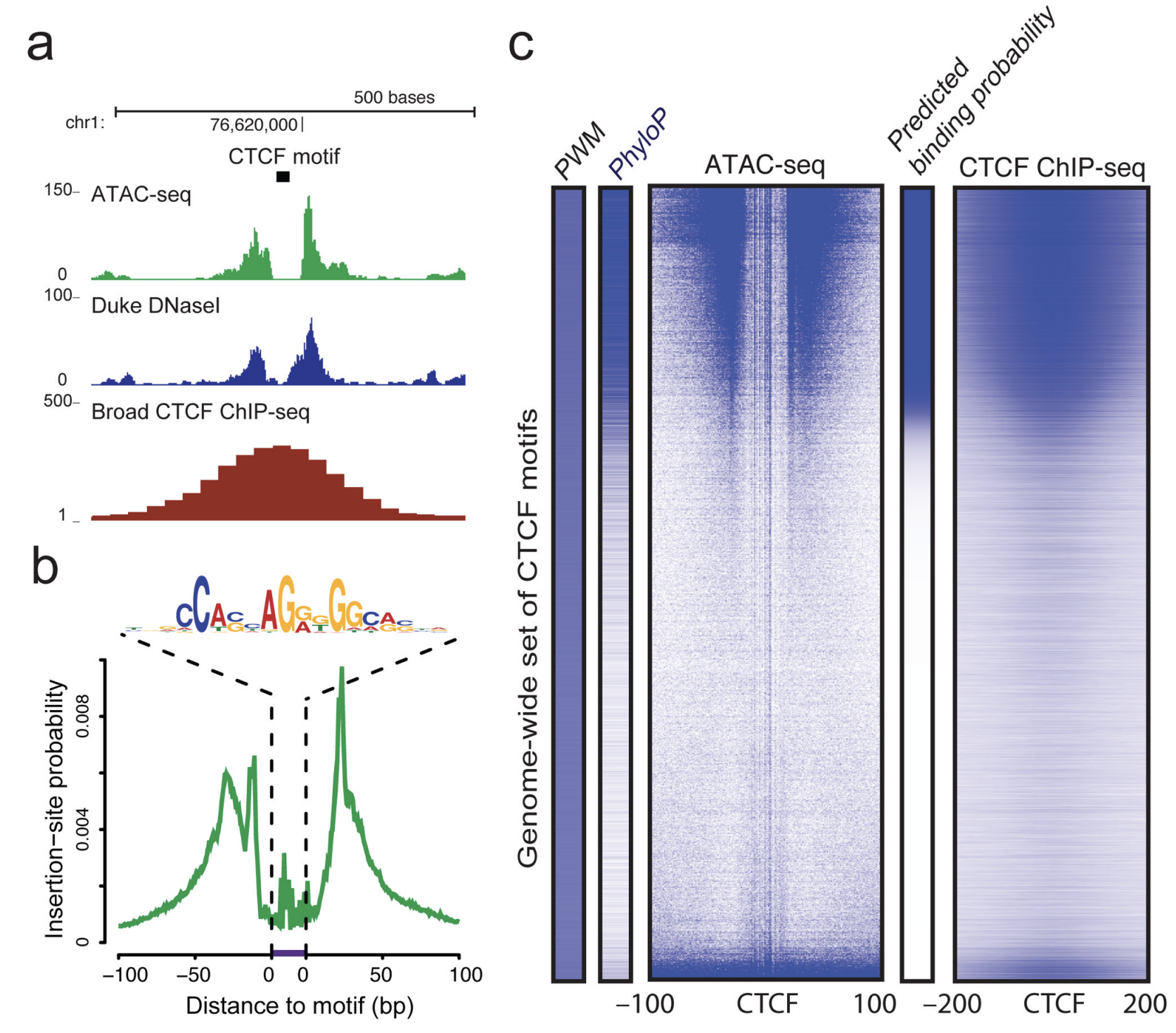
The 3 columns are. We will use the RNASeqPower R library by the same authors which implements a formula for the number of samples given the above parameters plus the technical ones such as the coefficient of variation and depth of coverage for each regulatory element. Overview of ATAC-seq Buenrostro et al 2015.
ATAC-seq captures all open chromatin regions not just those bound by a specific factor. If cells are limited 50000 cells is also acceptable. This requires a sample sheet which is identical to that required by the ChIP-seq and RNA-seq workflows see ChIP-seq for details.
ATAC-seq Assay for Transposase-Accessible Chromatin with high-throughput sequencing is a method for determining chromatin accessibility across the genome. Each Tool is its own Docker container that accomplishes a specific step in the analysis workflow. For each sample prepare a mixture of 560 μL of Buffer PB from Qiagen MinElute PCR Purification Kit with 10 μL of 3 M sodium acetate pH 52 in a labeled Eppendorf tube.
- Goal is to obtain 50 million genomic reads per sample minimum to assess open vs closed chromatin regions and 200 million genomic reads per sample to detect transcription factor binding sites. Sample Types Active Motifs services team is the only group routinely generating ATAC-Seq data from tissues. The genome browser tracks shown here are at the position of the FOXP3 gene which encodes a marker of post-mitotic neurons.
It utilizes a hyperactive Tn5 transposase to insert sequencing adapters into open chromatin regions. The peaks indicate open chromatin signal at the promoter of the FOXP3 gene. For simplicity we will focus on a particular project which had the highest sample purity.
We recommend preparing 100000 cells for ATAC-Seq. When analyzing Chromium sequencing data for a given sample you must combine the reads associated with all four of the oligonucleotides in the sample index. ATAC-Seq Pipeline Installation Building promoter regions for QC and downloading motifs Running the ATAC-Seq analysis pipeline for a single sample Plotting the key ATAC-Seq Quality Control metrics ATAC-Seq pipeline output files Differential peak calling Intersecting peaks with annotation tracks Plotting peak density along all chromosomes ATAC.
HemTools atac_seq -f fastqtsv Sample input format fastqtsv This is a tab-seperated-value format file. The sample processing workflow is similar to that of ChIP-seq. See our sample processing workflow for more details.
Sample Index Sets for Single Cell ATAC Specifications Last Modified on August 31 2021 Permalink Chromium sample indexes each comprise four oligonucleotides. Should be the same for all replicates. Sequencing Metrics Base Composition of Chromium Single Cell.
For proteins with a known sequence binding motif the included software can identify those motifs that are enriched in open chromatin on a cell-by-cell basis. Human and animal tissues including xenografts and human biopsies Primary cells including T and B cells FACS sorted cells Most rare cell populations. There are some small changes however and well discuss them here.
For ChIP-seq data we also provide strand cross-correlation metrics ie those attached pdf files. ATAC-Seq Kit was used to assess the genome-wide open chromatin state of mouse brain tissue samples. - Use 1 μl of each diluted library to measure DNA concentration by QuBit.
It is an unbiased genome-wide approach to look for epigenetic changes in a sample. In this tutorial we will start with the raw count matrix of read counts per sample columns and peak rows. 5 minute incubation at 72 C to allow extension of both strands.
We can perform the differential open chromatin analysis using the --sampleSheet option of the ATAC-seq workflow. SRA_ID of ATACseq_50k_Rep2 sample. 03 Setup Tools and Methods We will use both RBioconductor packages as well as other software packages for ATAC-seq analysis.
Single-End Config This is the config file format for single-ended data. The Bio-Rad ATAC-Seq Analysis Toolkit is a command line Tool that is packaged into Docker containers. 5 cycles of amplification PAUSE amplify a subset using qPCR to determine optimal cycle number 2.
Single Cell Data Highlights and Comparisons. Cd hindbrain_forebrainwork bcbio_nextgenpy confighindbrain_forebrainyaml -n 16 Parameters peakcaller. Single end sequencing for ATAC-seq is strongly discouraged for reasons mentioned below.
Macs2 bcbio just supports MACS2. Ad Simple and rapid tool for analyzing chromatin accessibility genome-wide. Single nuclei suspension prepared from fresh cryopreserved and flash frozen tissue or cell samples is needed for library prep.
Quality Control The quality metrics are provided in the html report. First of all the sample should be sequenced using the paired end PE protocol. Chromium Next GEM Single Cell ATAC v2.
Single cell ATAC-seq using 10x Chromium. Config File A config file is a tab separated text file that includes information regarding the name location and input of your experiment. Interpreting Cell Ranger ATAC Web Summary Files for Single Cell ATAC Assay.
The merge ID that will be used should your files be merged together. If you are interested in the basic Bioinformatics methods how ATAC-Seq data is aligned to the human reference genome quality controlled and processed for peak calling and peak annotation you can have a look at our ATAC-Seq pipeline. ATAC-seq overview Buenrostro et al 2015.
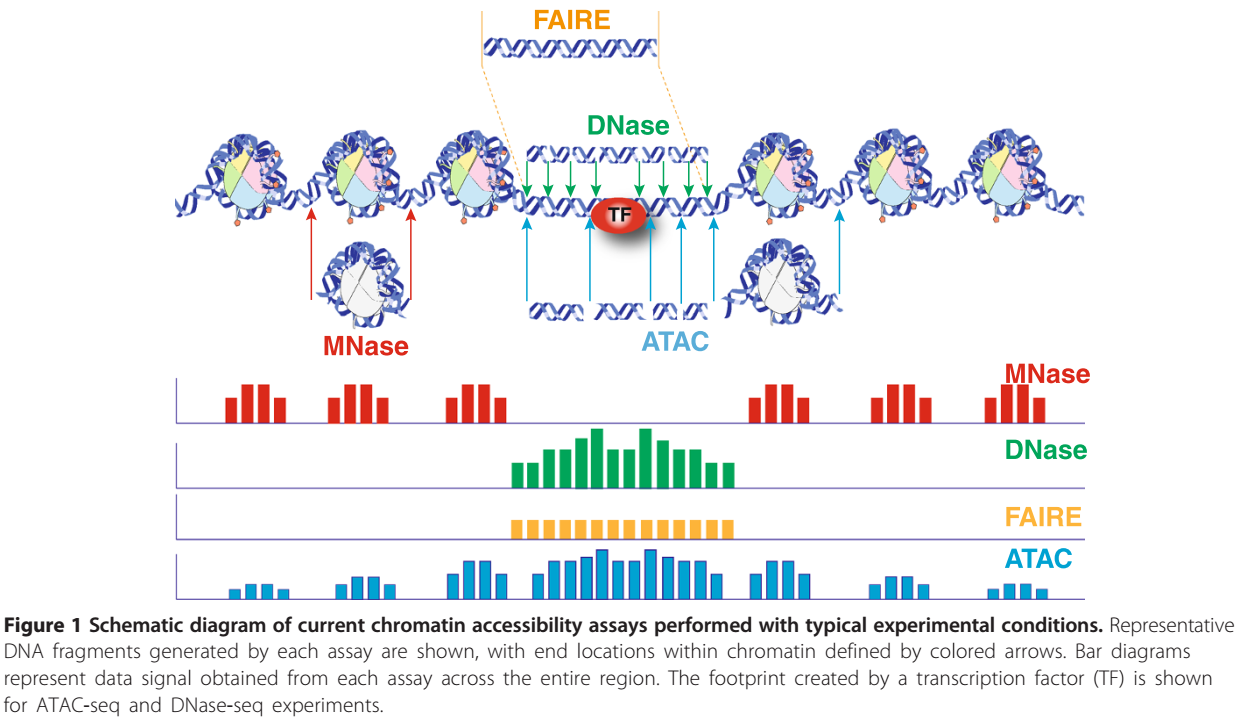
No comments for "Atac-seq Which Index to Use With One Sample"
Post a Comment